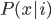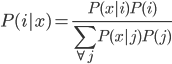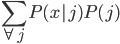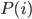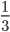Update: Pinterest now lets you disable Picked For You pins! Go to your profile, then press the settings button (a hexagonal bolt). Scroll down to "Home feed", then switch 'Show "Picked for you" Pins in your home feed' to 'No'.
Thank you all for your kind comments and bug fixing for this script. I hope it was useful.

It's no secret that I love Pinterest more than is really okay. They've started doing this thing where they show you "Picked for you" pins on your homepage, interspersed with your friends' pins. I quite like the curated pins they choose, but sometimes I like to just see what my friends have been up to.
You have the option to go through and deselect all of your boards, which I assume would prevent you from getting "Picked for you" pins at all. However, I wanted something a little less permanent, so I've made a bookmarklet.
The easiest way to use it is to drag the following link onto your bookmarks toolbar:
Remove Picked For You
Then, if you go to pinterest.com and click on your new bookmark, the "Picked for you" and "Promoted by" pins will have disappeared.
To keep the pins hidden, you have to click on the bookmark every time you load the page. However, there's a browser add-on for Firefox (or Iceweasel) called Greasemonkey, which lets you run little bits of javascript all the time. First, install Greasemonkey. Then, copy the text below:
// ==UserScript==
// @name Remove Picked For You
// @namespace bethmcmillan.com
// @include *
// @version 2
// @grant none
// ==/UserScript==
function hide_picked_for_you() {
divs=document.getElementsByClassName("creditFooter");
for(i=0;i<divs.length;i++)
{if (divs[i].innerHTML.indexOf('Picked for') != -1)
{divs[i].parentNode.parentNode.parentNode.style.visibility='hidden';}
}
divs=document.getElementsByClassName('creditName');
for(i=0;i<divs.length;i++)
{if (divs[i].innerHTML.indexOf('Promoted') != -1)
{divs[i].parentNode.parentNode.parentNode.parentNode.parentNode.parentNode.style.visibility='hidden';}
}
}
window.setInterval(function(){
hide_picked_for_you();
}, (5000));
Next, click on the Greasemonkey icon in your browser (usually in the top right), go to "New User Script", then "Use Script From Clipboard". Now, when you go to Pinterest and click on the Greasemonkey icon again, there should be the option "Remove Picked For You". If you tick it, you'll see that the pins disappear. For Google Chrome, there's an extension called Tampermonkey, which works in the same way.
If the script or bookmark aren't working for you, try reinstalling them from this page, as I might have updated the code. If this doesn't fix it, tell me in the comments. The latest code is always available on Github.