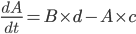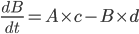We had the first meeting of our cardiac modelling journal club yesterday, and we looked at this paper by C. Frank Starmer from 2002.
The paper was investigating "Sanderson's paradox", whereby drugs intended to treat cardiac arrhythmia actually increased the incidence of sudden cardiac death in patients, even though they displayed what were thought to be anti-arrhythmic properties in single cells. (Incidentally, I think "paradox" is an overly dramatic word for "we discovered that we were wrong about something"). He was looking specifically at sodium blockers, because they were common anti-arrhythmic drugs.
He first describes a method for modelling drug block: it has been observed that sodium blockers are more effective when the heart is paced faster. This would suggest that the drug might bind only to the open ion channel. He formulated this as an extra gate on the sodium channel in addition to the h, m, and j gates.
He next talks about the refractory interval, which is the length of time a cell spends being unable to depolarise again after the initial stimulus. It was generally considered that increasing the refractory interval would be anti-arrhythmic, because it would make the cell less vulnerable to being stimulated by some random event.
So the thought was that you'd start with this:

And then your drug would change it to this:

Judging by the results of these drugs in humans, however, it looks like this isn't what happens on a larger scale. Starmer then looked at a group of cells arranged in a 1-dimensional cable, and talked about finding a different definition for the vulnerable period, based on the properties of the whole cable.
Usually when you apply an electrical stimulus (S1) (like you'd get during a heartbeat) to the end of a 1D cable, like so:

You get a wave propagating away from the stimulus, as expected:

This is all fine and normal, but what if you had a second stimulus (S2) outside of the usual time and place of stimulation?

You'd either get a wave propagating backwards along the cable:

or waves going in both directions:

In 1D, having an aberrant wave that propagates in one direction only is a pro-arrhythmic marker, because it could lead to a re-entrant arrhythmia in the whole heart. It's important, therefore, to work out what amount of drug block is going to cause unidirectional propagation and what amount bidirectional, and to redefine the "vulnerable period" as a property of the propagating wave. The vulnerable period depends on such things as the distance and time between S1 and S2, the speed of the wave, and the sodium channel availability.

If I have understood this paper correctly, the vulnerable region is the time between there being enough sodium channel availability at the location of S2 to create a backwards wave and there being enough availabiltity to create a forwards wave at the same location.
This was a really interesting thing to do, and I'm looking forward to having more journal club sessions, because it helped me to pick out the parts of the paper that I thought I understood but didn't, and brought together a lot of clever people in a room to explain them to me.