I wrote this for the Wellcome science writing prize. It didn't make the shortlist, so I can publish it here!
Enzymes are the machines of the biological world. In our bodies, they break down our food, fight off invading bacteria, turn sugar into energy, and perform all the processes that keep us alive. Honed by millions of years of evolution, they perform reactions at lightning speed - thousands of times faster than they would happen naturally. Chemists look on in envy as enzymes effortlessly do things that would take them millions of pounds of equipment, and often several years, to copy.
At home, we're all familiar with enzymes as the secret behind biological washing powders, which clean stains at low temperatures. Commercially, enzymes are used to recycle paper, as well as in septic tanks, and even in kitchens as meat tenderisers. And they do this all at room temperature, without help from strong chemicals.
Making enzymes that perform any reaction we want could revolutionise modern living. Imagine soaking your potatoes in an enzyme bath that takes the skins right off, no peeling necessary. We could make enzymes to break down oil spills into harmless substances, make new medicines that target specific cancers or infections and destroy them – or even make a magic spray that eats away the horrible sticky marks that price labels always leave on books, without destroying the cardboard covers (this one is a dream of mine).
So why haven't we done this? Well, enzymes need to be a specific shape in order to work at all. Unfortunately, enzymes are a type of protein, and we still don't know how to make proteins that fold into the shapes we want. A protein chain will always fold into the same shape, time and time again - but the forces behind this are, for the most part, a complete mystery. Until we know this, it's impossible to make artificial enzymes that are anything other than floppy, useless messes.
We can copy proteins from the natural world, stealing structural sections and active sites and mashing them together. But - if the enzyme produced is even functional - we're still limited to reactions that real enzymes already do catalyse. So, we're stuck in this situation, where we can't design our own enzymes, because we don't know how to make the structure. But what if we could side-step that problem altogether, and make evolution do the work for us?
Enter antibodies. Antibodies are the first line of defence your immune system puts up against invading germs. They're proteins as well, and they bind very tightly to specific parts of the bacteria or virus your body is fighting, signalling to the rest of the immune system to come in and destroy it. New antibodies are produced every time your immune system comes across a new pathogen. Antibodies are very specific – each antibody binds to one thing, and it binds that one thing very well indeed.
Scientists raise specific antibodies against a particular molecule by injecting it into a rabbit or a mouse. The animal's immune system then makes new antibodies against the unfamiliar molecule, which can be taken out and grown in a petri dish.
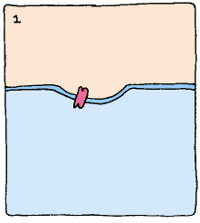
But what use is a protein that just binds tightly to something if you're trying to make an enzyme? Well, enzymes use a lot of different tools to make reactions happen. But if we take a step back and look at the whole picture, one common theme underpins them all: enzymes bind the most tightly to the intermediate stage of the reaction they supervise.
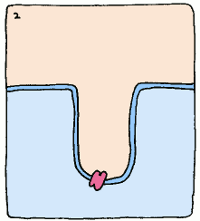
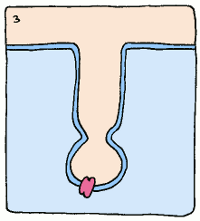
Take, for example, an enzyme that converts starch into sugar. It will bind to starch fairly well to start things off, but it will twist the starch molecule out of shape, because it really wants to bind to the intermediate. The intermediate's structure isn't quite starch, and it isn't quite sugar, but it's something in between. This twisting puts a strain on the starch, making the molecule react and form the intermediate. Intermediates are always unstable molecules, so then the intermediate will rapidly turn into the final product, which is sugar. Et voilà, the reaction has occurred.
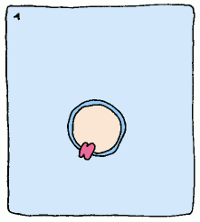
So if we can create a stable version of this intermediate and create an antibody which binds to it, we'll have a sturdy protein shape that catalyses a reaction the way we want it to – no assembly required. These new biological machines have been dubbed “abzymes”, and have already been used to target and inactivate the HIV virus.
Without having to unlock the secrets of protein structure, we've managed to make custom, functioning enzymes, just by relying on the immune system to do the work for us. Abzymes: lazy science at its best.