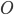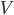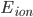For my research, I program using a collossal C++ software package called "Chaste". CHASTE stands in this case for "Cancer, Heart and Soft Tissue Environment", rather than its usual meaning of the sexual repression of people (especially women) all over the world. Interesting choice of acronym.
For my research, I program using a collossal C++ software package called "Chaste". CHASTE stands in this case for "Cancer, Heart and Soft Tissue Environment", rather than its usual meaning of the sexual repression of people (especially women) all over the world. Interesting choice of acronym.
Chaste's got lots and lots of bits that I don't use, and it is notoriously difficult to install on things, but it's also incredibly, incredibly useful. Over the last 10 years or so people have added all kinds of modules for biological modelling, especially cardiac stuff. Just a quick look at the class index gives you an idea of how many tools there are to use.
There's an easy Chaste installer for Ubuntu, but I'm a Debian girl myself, and I've had terrible trouble trying to use the generic instructions, because there are a lot of dependencies, and each dependency has different versions in different Linux distributions.
In the end, I decided to side-step the issue entirely. There's a tool called Debootstrap, which creates an entire Ubuntu installation in a single folder, so I can be using my Debian laptop and have Ubuntu open in a terminal without too much bother.
So first we make a Chaste directory and put Ubuntu into it using debootstrap:
mkdir Chaste sudo debootstrap utopic Chaste http://mirrors.kernel.org/ubuntu/dists/utopic/
Next, we jump into the Ubuntu installation by changing our root:
sudo chroot Chaste
First off for some reason you need to tell the computer where you are.
locale-gen en_GB en_GB.UTF-8
Then there are a few useful things that I like to have around; "nano", which is a text editor, "aptitude" which is a front-end for the apt-get installer, and "man" which are the manual pages for command line tools.
apt-get install nano aptitude man
Now you need to add some more repositories so that you have all of the packages available that you need. Open up your sources file:
nano /etc/apt/sources.list
Delete the first line using Ctrl-K, and then copy in the following:
deb http://archive.ubuntu.com/ubuntu utopic main universe multiverse deb http://archive.canonical.com/ubuntu utopic partner deb http://archive.ubuntu.com/ubuntu utopic-backports main restricted universe multiverse deb-src http://us.archive.ubuntu.com/ubuntu/ utopic universe main multiverse deb http://us.archive.ubuntu.com/ubuntu/ utopic-updates universe multiverse deb-src http://us.archive.ubuntu.com/ubuntu/ utopic-updates universe multiverse deb http://www.cs.ox.ac.uk/chaste/ubuntu utopic/
Then press Ctrl-O to save and Ctrl-X to exit. Next, we add the Chaste license key, and update the package lists.
apt-key adv --recv-keys --keyserver hkp://keyserver.ubuntu.com:80 422C4D99 apt-get update
The proc filesystem needs to be mounted before we can continue:
mount -t proc proc /proc
After this point, I am just following the default install guide. This next part took forever, I got quite bored. Here is a collection of margin scribbles from books.
apt-get install --install-recommends chaste-dependencies apt-get install `dpkg -s chaste-dependencies | egrep "^Suggests" | cut -d "," -f 1-111 --output-delimiter " " | cut -d ":" -f 2`
Then checkout the code! I put it into a folder called "Chaste" in the root directory, but if you want to put it somewhere else (/home/scratch/Chaste/ for example) just create the directory you need first and cd into it.
svn --username anonymous checkout https://chaste.cs.ox.ac.uk/svn/chaste/trunk/ Chaste cd Chaste scons
Then all of the tests should start building!