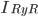I gave a presentation yesterday to the Cardiac group here at Oxford about my summer project and my plans for my PhD. It was, quite frankly, terrifying, which is fairly typical of all the presentations to group meetings that I've ever given. Talking to people who are experts in your subject about what you're doing is quite a nerve-wracking experience, no matter how prepared you feel you are.
Nonetheless, it was a really good opportunity to get feedback and advice on my results and on my next steps. I have been a bit concerned that because my simulations involve changing ionic channel conductances to up to 50x their usual level, my results wouldn't be physiologically relevant. Blanca brought up the point that a healthy heart isn't likely to have an arrhythmia, so ion channels that aren't operating outside normal limits aren't particularly relevant to my research.
The next time I present my research plan (which will be in 2 weeks' time at my pre-PhD viva) I think I'll include a flow chart of the process, because most of the questions asked afterwards were along the lines of "Sorry, what?", which is non-optimal. I'm also determined to start using LaTeX for my slides - LibreOffice, while wonderful, refuses to display .eps images, for some reason. I'd like to be able to send data from my simulations straight through my Matlab scripts and into a presentation, without tedious faffing with the Gimp or ImageMagick and trying to avoid horrible pixellation.
I included appendices after the main presentation to answer questions that people might ask, which was an idea I stole off one of the students in the Computational Biology group. Nobody did ask the questions, in the end, but it was nice to have the extra information there just in case.
The one question I wasn't prepared for was "Why did you choose these particular models?" I chose them because they have been reported in the literature to show both DADs and EADs, but when I was asked the question I froze and stammered something about those models being the ones that didn't break my program (which is also true, but less relevant). That wouldn't have been appropriate for an appendix, but I'll make sure to write it on my cue cards the next time, so I have the information at my fingertips and don't have to rely on my memory — which is a notorious cad and bounder, and often deserts me at a moment's notice.
You can download my presentation from here if you are particularly interested.


 The EAD begins at around 0.3 seconds.
The EAD begins at around 0.3 seconds. The DAD begins at around 2 seconds. The sarcoplasmic reticulum calcium release current is labelled as
The DAD begins at around 2 seconds. The sarcoplasmic reticulum calcium release current is labelled as